FcLigand software:
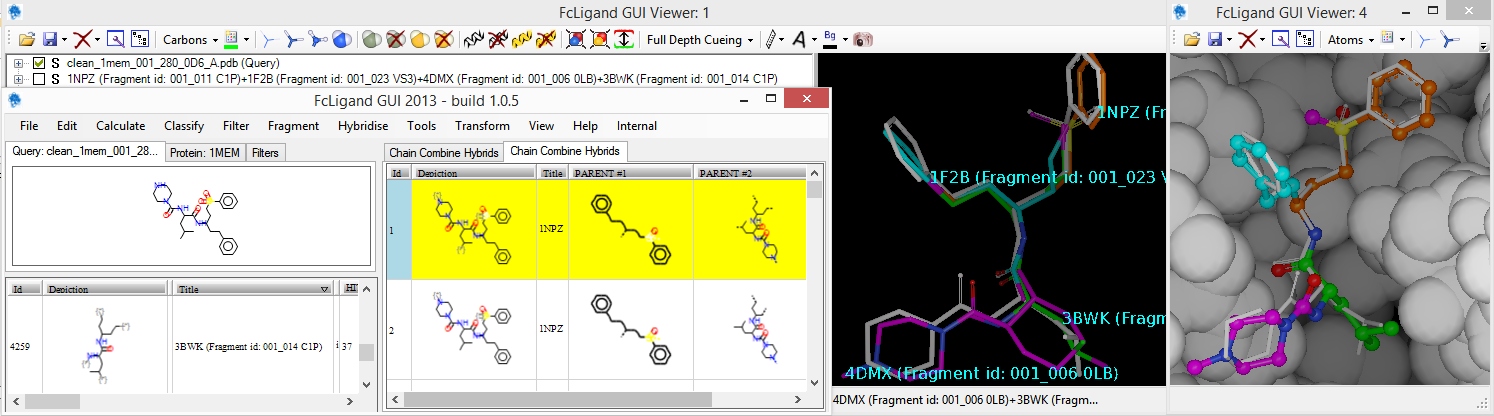
FcLigand, a powerful cheminformatic software for Lead Generation and Lead Optimisation has been designed to take advantage of biostructural and chemical library information. Protein Data Bank is now having more than 170000 entries with protein(s)-ligand(s) complexes (Nov 2020), it’s an attractive source of innovation when mined with a software such as MED-SuMo from MEDIT to 3D compare and superpose protein/ligands in binding sites or subpockets. From such pool of ligand/fragment alignment, FcLigand provides a rich environment to deconvolute ligands in smaller entities, or to 3D combine fragments and ligands in larger molecules with a chemical diversity few times higher than the PDB. FcLigand is essential for FBDD (Fragment-Based Drug Design). FcLigand run under Microsoft Windows operating system.
Click here to download FcLigand PDF brochure
FcBioisostere software:
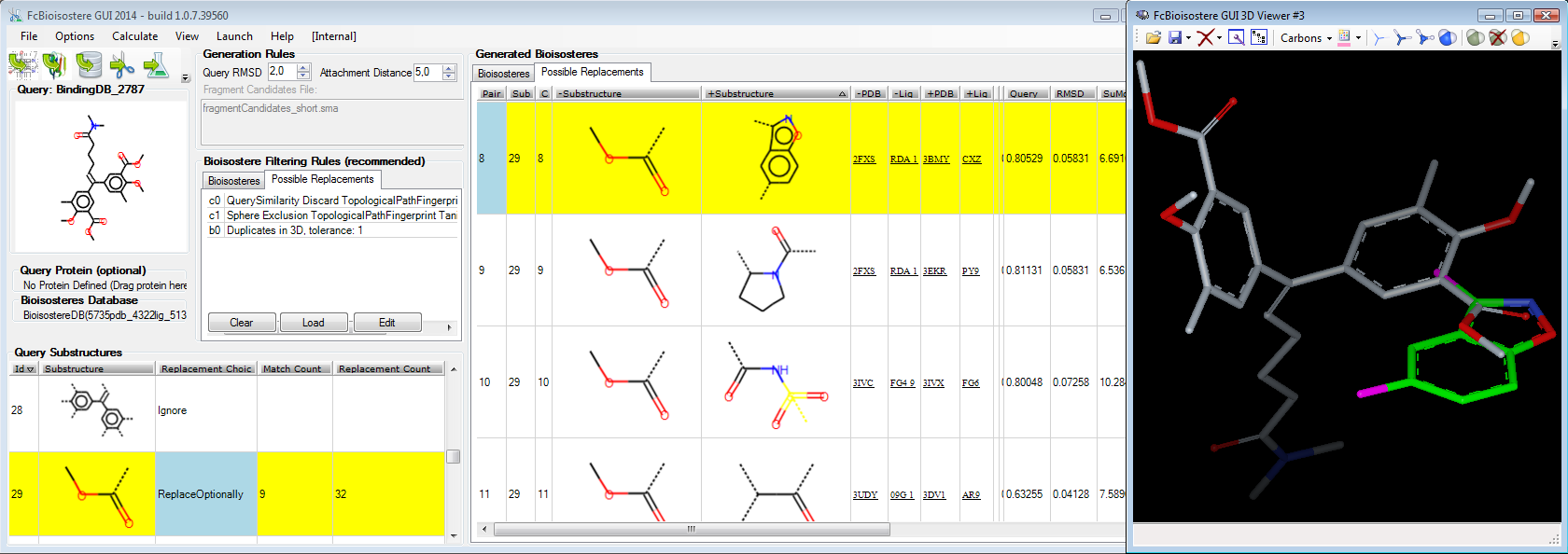
FcBioisostere provides access to 3D bioisosteric replacement onto your molecule of interest to find chemical groups having similar 3D biological interactions. While maintaining target potencies, it helps Chemists to optimize additional properties in pharmacokinetics and metabolic response, and/or to escape from existing patents by selecting alternative equivalent chemical groups.
FcBioisostere is a 2 steps application: First, users can automatically build a database of 3D bioisosteric fragment pairs from :
-1a- an input set of pre-aligned ligands (3D SD files) where pairs of 3D overlapping fragments are detected and stored ; or
-1b- by driving MED-SuMo MEDIT SA software to extract from the PDB files (Protein Data Bank, ~150000 protein structures) and/or your in-house biostructural data all local protein-ligand similar superpositions (defined by charges, Hbond, hydrophobic, aromatic, … interactions); or
-1c- alternatively we provide pre-computed databases of bioisostere pairs built upon the whole PDB prepared by Felix Concordia experts.
Second, the input molecule of interest is deconvoluted in fragments (6 methods). Once fragments to be replaced are selected, all fitting replacement pairs of fragment are retrieved from the database. Different methods to merge/fuse those fragments into the input molecule are provided to generate full bioisosteres. 1D/2D/3D chemical and protein scoring filters help to sort/focus on the best bioisostere candidates. FcBioisostere run under Microsoft Windows operating system.
Click here to download FcBioisostere PDF brochure
FcRebuilder software:
 FcRebuilder tries to predict ligand 3D bound conformation in binding site(s) by mining the PDB biostructural knowledge (Protein Data Bank). From pool of aligned PDB subpocket-ligand structures, FcRebuilder is extracting substructures corresponding to the ligand to predict and then 3D-combining iteratively these fragments to rebuild as much of the 2D input ligand in a bound conformation. We shown on a dataset of 2241 ligands in single PDB entry that in 52% of the case this public database was hosting enough 3D knowledge to rebuild at least 80% of 3D bound coordinates. FcRebuilder is definitely bringing a new approach in Docking methodologies by taking advantage of all existing biostructural knowledge.
FcRebuilder tries to predict ligand 3D bound conformation in binding site(s) by mining the PDB biostructural knowledge (Protein Data Bank). From pool of aligned PDB subpocket-ligand structures, FcRebuilder is extracting substructures corresponding to the ligand to predict and then 3D-combining iteratively these fragments to rebuild as much of the 2D input ligand in a bound conformation. We shown on a dataset of 2241 ligands in single PDB entry that in 52% of the case this public database was hosting enough 3D knowledge to rebuild at least 80% of 3D bound coordinates. FcRebuilder is definitely bringing a new approach in Docking methodologies by taking advantage of all existing biostructural knowledge.
FcRebuilder includes a Heat-Map mode to manage rebuilding on multiple targets, an optional connexion to FcBioisostere database to enrich rebuilding performances with PDB-based bioisosteric pairs, and is optimized for multicore processor architecture. The FcRebuilder implementation in FcLigand provides further tools to filter, cluster, and score all 3D bound poses.
Click here to download FcRebuilder PDF brochure
FcCutlass software:
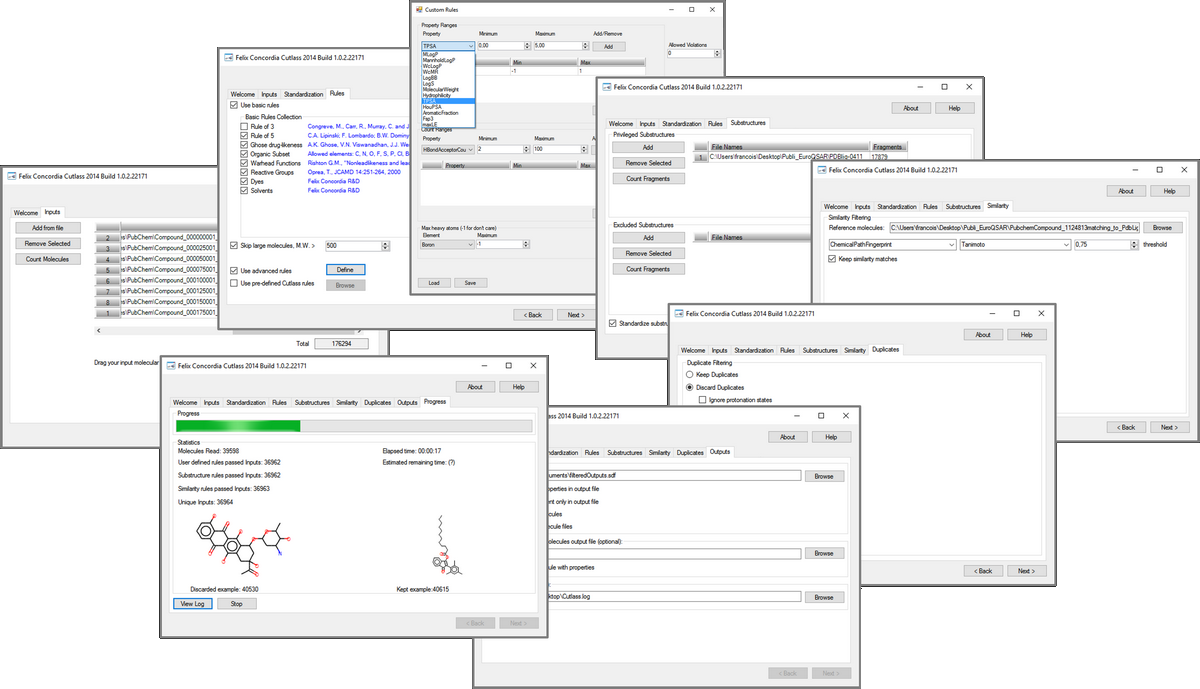
FcCutlass is a graphical tool for filtering molecules and fragments. It allows to manage several source of chemical libraries in order to extract the best subset of data according to (a) 1D-2D-3D physico-chemical descriptors thresholds, (b) existing sub or super structures, (c) undesirable chemistry, (d) protein-ligand scoring function when your input molecules are already 3D superposed in a protein binding site, (e) chemical diversity by clustering molecules, (f) exiting 2D or 2D/3D duplicates molecules. It was built upon the C2P-API. FcCutlass run under Microsoft Windows operating system.
MEDIT’s partner products:
MED-SuMo MEDIT software compares and superposes biostructures by detecting 3D chemical features on protein surface arranged with similar orientations, inter-feature distances and inter-triplet angles (neighboring chemical features are defining triplets). A GUI provides advanced searching capabilities onto biostructures of the Protein Data Bank database to define subpocket-to-full surface queries. The interactive result analysis can discriminate unique to very frequent intermolecular features within a protein family or across the full PDB, and export all 3D protein-ligand generated alignments to other software for further drug design applications.
