DOWNLOAD
C2P-API Material:
The Chemo-Proteomic Advance Programming Interface provides a rich set of documented methods for drug design. We are working hard to provide very soon an access to the source code and to the documentation.
Other R&D Material:
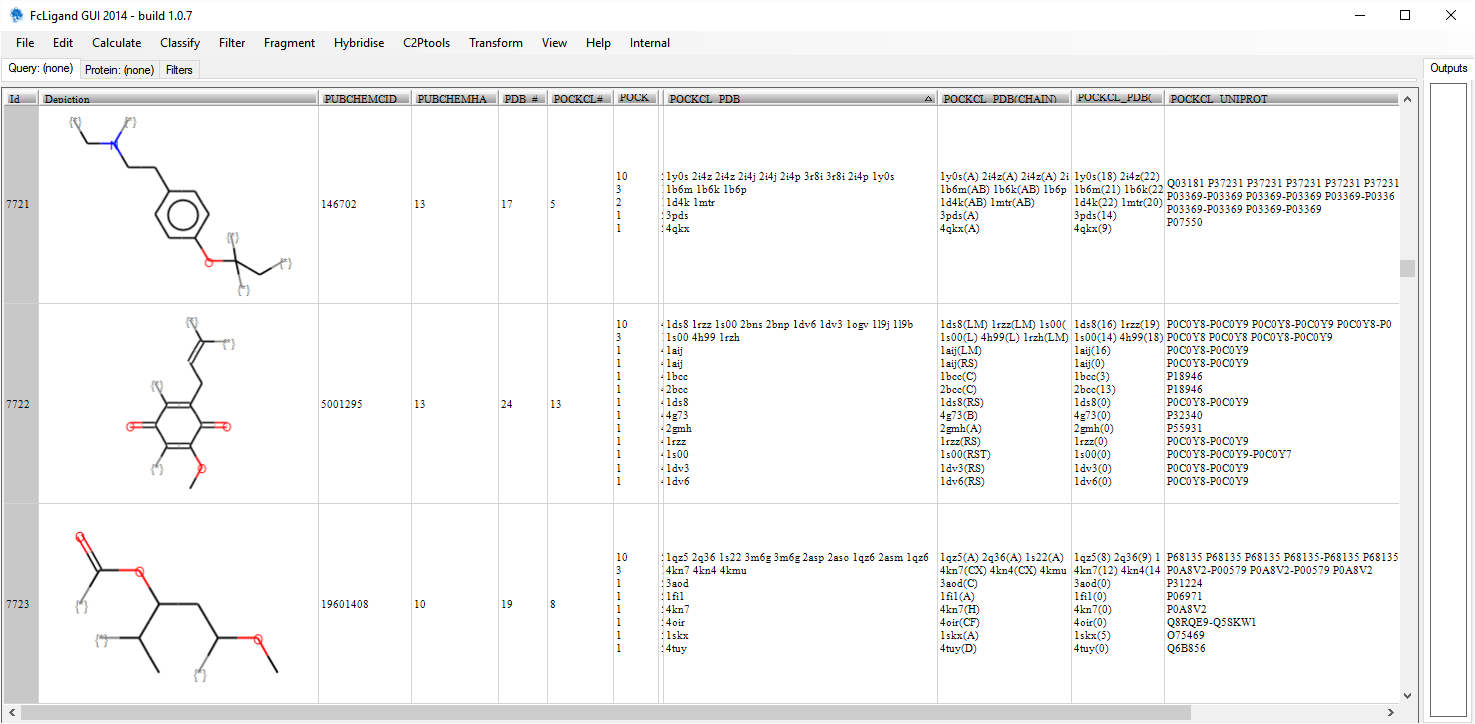
- PDB analysis of subpocket similarities per 2D fragment structures: this file contains a set of 2D fragment structures with additional annotations: the Pubchem CID, the number of heavy atoms, the number of PDB codes having a ligand that is a molecular superstructure of the 2D fragment, the number of subpocket clusters, the size of each subpocket cluster (split by a carriage return), the PDB code with the protein chain identifier(s) defining the binding site, the number of SCF defining the subpocket for each PDB code, the list of Uniprot codes per cluster, the list of Pfam families per cluster, the list of Pfam distribution and last the Uniprot distribution (Pfam and Uniprot were annotated only when available from the PDB). Click please on the image below to download the SDF file.
- List of PDB ligands uniquely represented in the PDB structures (as Oct 13th 2014) : this file was set to evaluate the feasibility of predicting 3D ligand atomic positions in binding sites based on PDB-derived information (submitted paper). The following criteria were used to define a list of reference ligands: (a) X-ray resolution under 2.5 Å, (b) ligand MW between 350 and 550 Da, (c) ligand in unique position in the asymmetric cell to avoid cases where the ligand would be reconstructed from another available bound conformation in the PDB file, (d) Ligand in unique entry across the PDB. We obtained a list of 2,309 macromolecule/ligand complexes. Click here to download this XLS file
