Proof Of Concepts to annotate protein surfaces and generate drug candidates:
Since the beginning of the COVID-19 pandemic crisis, the scientific community has generated an amazing amount of data especially in structural biology with more proteins together with cocrystallised ligands of fragment probes. We ran our 3D intermolecular surface miner protocol in to first superpose the PDB materials having similar molecular surface, then combined in FcLigand the superpose ligand/fragment materials into protein pockets, and finally annotated the generated molecules by matching to PDB/Pubchem/ChEMBL repositories.
On Spike protein that project from SARS-CoV-2 surface (pdb code 7JV4), we compared the full 3D surface to retrieve PDB proteins showing similar local network of 3D interactions. The resulting superposition highlights epitopes positions on the RBD Rigid Body Domains targeted by actual vaccines.
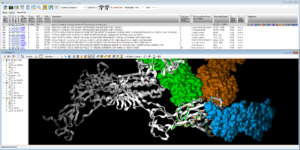 Fig. Epitope detection on SARS-CoV-2 Spike protein: spreadsheet on top summarizes 19 non-SARS-CoV-2 proteins that interact with Spike protein ; 3D viewer displays in white the 7JV4 query backbone and 3 hits in ball rendering, n°494 (7LM9 in blue), n°604 (2DD8 in orange), and n°848 (7JN5 in green vert) ; only antibody part of hits are displayed, network of 3D shared structural chemical features (Hbond donor/acceptor, charge, hydrophobicity, aromatic, …) for the 3 hits highlights putative epitope surfaces.
Fig. Epitope detection on SARS-CoV-2 Spike protein: spreadsheet on top summarizes 19 non-SARS-CoV-2 proteins that interact with Spike protein ; 3D viewer displays in white the 7JV4 query backbone and 3 hits in ball rendering, n°494 (7LM9 in blue), n°604 (2DD8 in orange), and n°848 (7JN5 in green vert) ; only antibody part of hits are displayed, network of 3D shared structural chemical features (Hbond donor/acceptor, charge, hydrophobicity, aromatic, …) for the 3 hits highlights putative epitope surfaces.
Onto NSP 5 Main Protease (pdb code 6Y2F), we collected 209 PDB ligands having 3D similar interactions with the query and no strong steric clashes, then 3D-combined by detecting the Maximal Common Substructure to obtain 2320 hybrids ; 24 were 2D-matching to PDB ligands, 92 to Pubchem molecules, 2 to ChEMBL molecules.
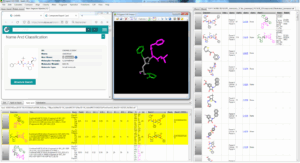 Fig. FcLigand 3D-hybrisdisation of ligand structures from 209 PDB complexes where 3D similar interaction surfaces were detected and superposed onto the 6Y2F binding site ; the displayed hybrid corresponds to CHEMBL 213054 by 3D hybridisation of 3SZN, 3AVZ and 7C8T co-crystallised ligands (shared detected scaffold is in gray)
Fig. FcLigand 3D-hybrisdisation of ligand structures from 209 PDB complexes where 3D similar interaction surfaces were detected and superposed onto the 6Y2F binding site ; the displayed hybrid corresponds to CHEMBL 213054 by 3D hybridisation of 3SZN, 3AVZ and 7C8T co-crystallised ligands (shared detected scaffold is in gray)
Additionally, we used FcRebuilder on MPro to dock 212 ChEMBL active molecules on SARS-CoV-2 targets ; our software first retrieved 900 protein-ligand structures with a 3D interaction surface similar to our selected 6Y2F target. These superposed ligands were then used to 3D-rebuild the ChEMBL subset into the binding site; the predicted poses positioned at least 90% of the structure.
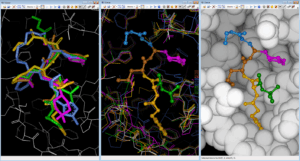 Fig. Rebuilding/Docking ChEMBL212080 into 6Y2F MPro with FcRebuilder software ; left view shows after 3D superposition into binding site five cocrystallised ligands from 7JW8 in green; 7M00 in magenta, 2A5K in yellow, 7LBN in brown, and 6LKA in blue ; middle view shows the fragment contribution to rebuild ChEMBL 212080 molecule ; right view displays the pause in the MPro target.
Fig. Rebuilding/Docking ChEMBL212080 into 6Y2F MPro with FcRebuilder software ; left view shows after 3D superposition into binding site five cocrystallised ligands from 7JW8 in green; 7M00 in magenta, 2A5K in yellow, 7LBN in brown, and 6LKA in blue ; middle view shows the fragment contribution to rebuild ChEMBL 212080 molecule ; right view displays the pause in the MPro target.
These 3 POCs highlight the benefit of our PDB based drug design protocols with our software technology
