FcBioisostere new graphical features to better explore bioisosteric PDB based pairs:
This year we extanded the FcBioisostere software to better sort the precalculated 3D bioisosteric pairs extracted by our application from the PDB Protein Data Bank. Same topological pairs are now grouped together to help users assess a statistical suitability of the proposed fragment replacement onto their ligand of interest. Furthermore, Structural Chemical Features (Hbond donor/acceptor, charge, hydrophobicity, …) and protein of proposed bioisosteric pair can be displayed alongside the input ligand.
These and other user enhancements are now available in the latest FcBioisostere release.
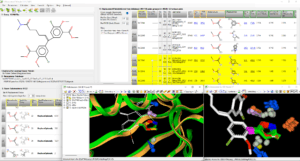
Fig.: Replacement of hydrolysable methyl ester chemical moiety on ADAMs ligand query (pdb code 3SI9) with FcBioisostere ; on the left, upper view shows the input 2D or 3D query, lower table shows all query substructures to select the area to replace ; on the right, upper table shows all found replacement pairs, the selected pair is yellow was detected in 121 PDB pairs, lower view shows first the query ligand in white with the protein-fragment pairs in green and orange, then on the right, some of the 121 pairs of methyl-ester by 1,2-oxazole replacement.
