Designing protocols with a workflow user interface:
In order to faster design new protocols for users in our C2P Chemo-Proteomic Platform, we investigated technical solutions to provide a sketcher to users to design predictive protocols similar to a workflow.
FcPipeline our new module under development provides a full set of input/calculation/output nodes that can be chained together to define any chemoproteomic protocols. The user interface dynamically displays the number of processed molecules at each stage of the workflow. The processing of ligand and protein flows is managed using iterator technology, which allows for immediate results for a given protocol, enabling users to verify the protocol’s validity without having to wait for the entire calculation to finish.
Any of our C2P API methods can be integrated as a separate node, with parallel processing enabled for that node if needed to improve performance. We plan to include all our chemo-proteomics C# methods in a future release of FcPipeline.
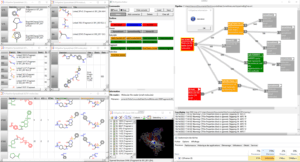 Fig.: FcPipeline protocol for fragmenting and 3D-hybridizing two sets of pre-superimposed ligands; red nodes represent input readers; orange and green nodes represent cheminformatics methods; gray nodes are viewers (all displayed on the left side of the view, with the 3D hybrid structures shown below, using red or green substructures to indicate the source ligand)
Fig.: FcPipeline protocol for fragmenting and 3D-hybridizing two sets of pre-superimposed ligands; red nodes represent input readers; orange and green nodes represent cheminformatics methods; gray nodes are viewers (all displayed on the left side of the view, with the 3D hybrid structures shown below, using red or green substructures to indicate the source ligand)
